Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
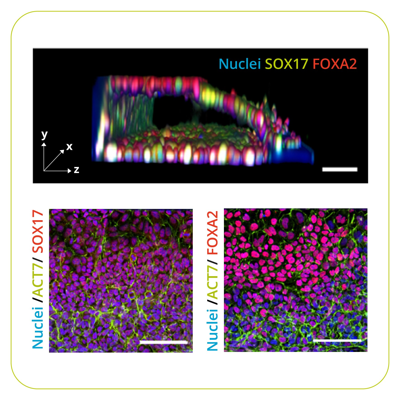
Millions of people worldwide experience pain and stress due to intestinalintestine-on-a-chip inflammation, emphasizing the need for the development of diagnostic strategies and therapeutic interventions. However, without suitable research models for studying intestinal functions, progress in research and drug discovery is difficult. The OrganoPlate® was developed to fill this gap and provides a robust, flexible platform that is amenable to drug screening. Recent adaptations1,2 of a previously established intestine-on-a-chip model3 has paved the way for new opportunities in high-throughput drug discovery and research related to intestinal inflammation.The OrganoPlate is a microfluidic platform that facilitates research across a wide range of physiological processes, including hepatic, renal, and intestinal function. A recent publication, by scientists at MIMETAS and international collaborators at the University of Sheffield describing the Direct On-Chip Differentiation of Intestinal Tubules from Induced Pluripotent Stem Cells (IPSCs) demonstrates improvements on the existing model, and represents significant progress towards a versatile, more complex platform for future drug discovery efforts.The need for advanced in vitro modelsInflammatory bowel disease (IBD) is a collective term used to describe a range of disorders involving intestinal inflammation and is estimated to affect more than 6.8 million people globally.4,5 The individual burden of IBD can be significant; those affected commonly experience chronic pain and stress, with symptoms ranging from mild to highly debilitating. Despite the clear need for preventative and therapeutic agents, such solutions are lacking – largely because the underlying mechanisms of intestinal inflammation remain poorly understood. In order to facilitate studies designed to explore intestinal inflammation and potential therapies, there is a need for research models that are:Robust and reproducibleAmenable to screeningSufficiently complexFlexiblePersonalizedIn recent years, major efforts have been directed towards the development of in vitro models of intestinal physiology: three-dimensional cultures, organoids, and microfluidics. Each of these provides important biologically relevant features, yet they are typically not suited for high-throughput screening. The OrganoPlate platform helps to fill this gap as it allows for the culturing of leak-tight, polarized epithelial gut tubules on a standardized platform that is compatible with high-throughput screening equipment.6 These features make it a valuable tool in drug discovery settings, as key aspects of IBD pathogenesis,e.g. increased cytokine production and the loss of barrier integrity can be recapitulated.6As a result, the OrganoPlate has been selected by several pharmaceutical companies to support studies into the mechanisms behind IBD, and diarrhea toxicity observed during a Phase 1 clinical trial.6,7 As that model was based on a single cell line, however, MIMETAS saw room for improvement and recognized the need to expand the OrganoPlate’s capacity for mimicking biological complexity.Towards patient-specific organoid tubules for personalized medicine approachesThere is no “one-size-fits-all” approach to the treatment of IBD, which is unsurprising given the substantial variation in phenotype and disease course that exists across individuals.8 As with many other conditions, a more personalized approach to the diagnosis and treatment of intestinal inflammation could improve clinical outcomes, as decisions could be based on a deeper understanding of an individual’s specific disease mechanisms. Such an approach has been made possible by technological advances, such as those in genomics and proteomics, but its potential is yet to be realized. For personalized medicine to become a reality, in vitro patient-specific models are needed to mimic intestinal diseases.With this goal in mind, MIMETAS' scientists and collaborators at the University of Sheffield developed a protocol for the directed differentiation of iPSCs into three-dimensional gut-like tubules in the OrganoPlate. An assessment of stem cell and adult intestinal gene expression markers revealed stepwise differentiation through definitive endoderm, hindgut, and intestinal stages, with barrier function and apical-basal polarity established over the course of two weeks. The iPSC-derived tubules were challenged with a cytokine cocktail to determine their susceptibility to inflammatory stimuli. Following exposure to tumor necrosis factor-α, interleukin-1β, and interferon-γ, an increase in gene expression of IL-6, IL-8, and CCL20 was observed in the tubules, indicating a significant inflammatory response.Completed in only 14 days, the protocol provides an efficient approach for personalized intestinal model development, without the need for an intermediary organoid step. Unlike organoid models, this approach enables straightforward access to both the apical and basal sides of the epithelium – a crucial consideration for drug development applications.
Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

Enhanced patient-specific intestine-on-a-chip model for disease modeling and drug discovery
This article describes advances in OrganoPlate® intestine-on-a-chip models—highlighting microfluidic platforms for high-throughput IBD research and new 14-day iPSC-derived gut tubules for personalized inflammation studies.

