What is 3D Cell Culture?
The function of a tissue is determined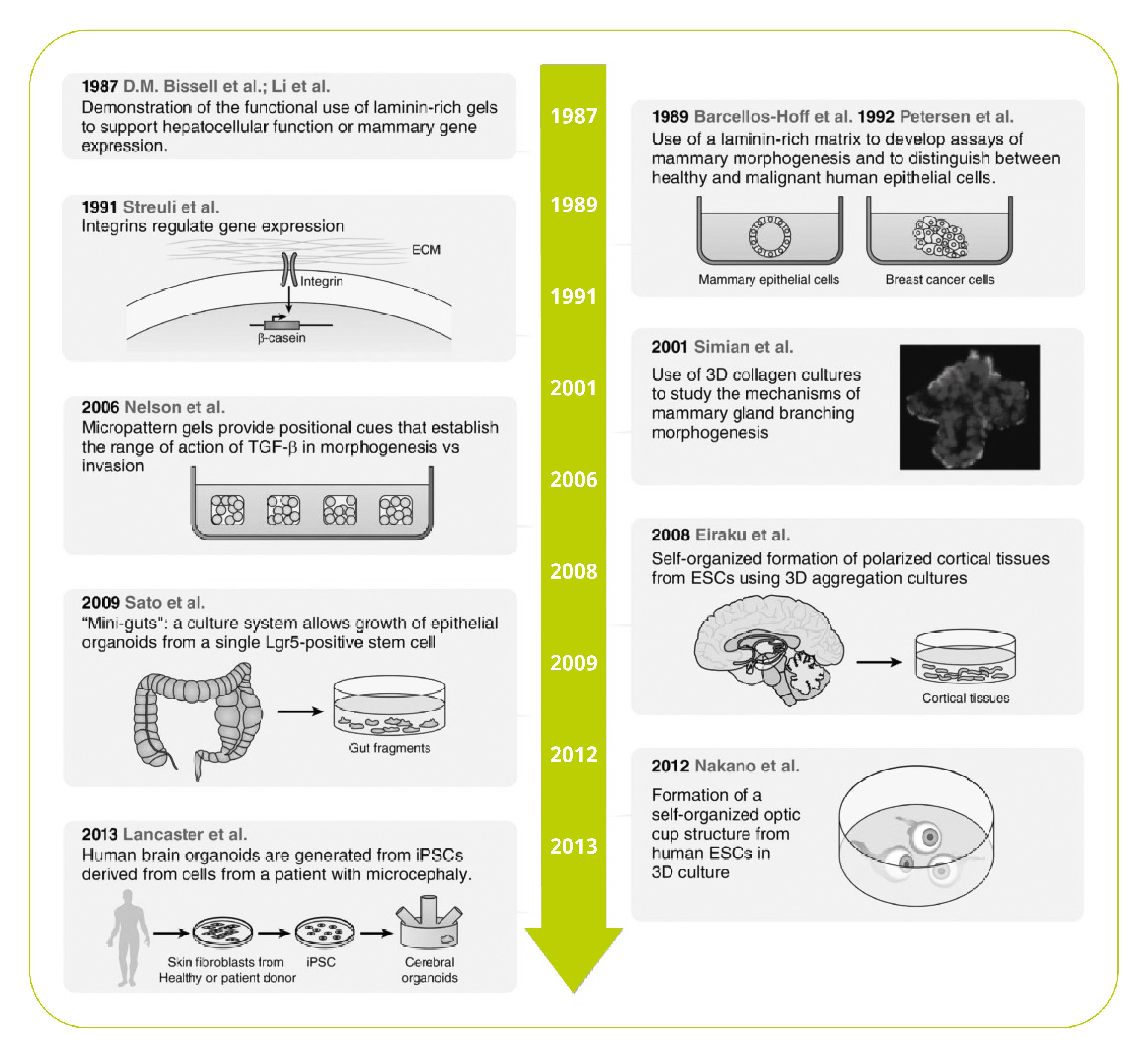 by both its cellular and non-cellular composition. The more closely a cell culture system can replicate those conditions, the more closely those cells will replicate the behaviors and responses of cells in the body. This makes 3D cell cultures appealing.
by both its cellular and non-cellular composition. The more closely a cell culture system can replicate those conditions, the more closely those cells will replicate the behaviors and responses of cells in the body. This makes 3D cell cultures appealing.
Cells in vivo experience a complex, three-dimensional environment that exposes them to circulating molecules, neighboring cells, and the extracellular matrix (ECM). With these environmental cues in mind, biologists have developed more physiologically accurate methods for culturing cells and tissues, including new media formulations and co-culture techniques. Back in the early days of cell culture, it was all we could do to keep cells alive and growing. Now, we have more ambitious goals – keeping cells alive, growing, and behaving just as they do in the body.
3D cell culture represents yet another step towards this goal by focusing on mimicking normal cell-matrix and cell-cell interactions. Along with microfluidic or organ-on-a-chip systems, 3D cell culture offers researchers an unparalleled ability to recreate physiological compositions and
spatial arrangements of cells in vitro.
A Brief History of 3D Cell Culture
We think we’re made of cells, but they are far from the whole picture. In fact, by mass, the most abundant proteins in our bodies are ECM proteins – the nonliving material outside of our cells that provides a physical scaffold, barrier, and mechanical and biological signaling.
However, that last role was underappreciated until the isolation of ECM proteins in the 1960s and 70s. These newly available materials led to compelling evidence that the physical cellular environment is far from inert, but acts as a biological and mechanical signal to regulate cell-type specific function, differentiation, and morphology1.
Since then, remarkable progress has been made in 3D cell culture, including organoids, miniature organ-like structures that self-organize in 3D culture. Additional advances in technology, including microfluidics and engineered synthetic ECM hydrogels, have furthered our understanding of the mechanical and microenvironmental cues that govern cell behavior2,3.
Why 3D Cell Culture?
2D vs. 3D Cell Culture
Simple, inexpensive, and reproducible, traditional or 2D cell culture is the mainstay of biological research. Meanwhile, 3D cell culture systems feature increased complexity for increased faithfulness to the in vivo environment. The primary changes to cells in 3D are rather obvious – shape, motility, and polarity are all affected – but can have important secondary, less-obvious tissue-specific consequences.
| 2D | 3D |
| High stiffness surfaces provide supraphysiological mechanical signals | Tunable, relatively low stiffness environment closer to that of tissues |
| Continuous, flat surface available for unencumbered adhesion, spreading, and migration | Nano- and micro-scale 3D surfaces provided by ECM fibers and matrix porosity guide and hinder cell motility |
| Soluble gradients absent without microfluidics | Gradients of soluble factors, nutrients, and oxygen based on diffusion through gel or cell aggregates |
| 2D geometry constrains morphogenesis and cell-substrate interactions dominate | Free to self-organize in 3D. In multicellular structures, cell-cell interactions dominate. |
| Automatic apical-basal polarization | Embedded cells generate apical-basal polarity on their own |
Adhesive, topographical mechanical and soluble cues in 2D and 3D4
A lot changes when a cell moves from 2D to 3D culture, but the most obvious is the mechanical environment. The stiffness of glass or plastic is multiple orders of magnitude higher than that seen in soft tissues, which have a consistency like Jell-O or cream cheese. Stiffness directly affects cell adhesion, spreading, migration, and differentiation. The mechanics of 3D cell culture, on the other hand, are quite different. ECM gels feature lower stiffnesses, local structures like collagen fibers, and pores that encumber migration and invasion. ECM proteins are also readily remodeled by the cells themselves by fiber bundling and degradation. Together, these can lead to reduced proliferation, altered migration, smaller and more distributed focal adhesions, and directional cell elongation along fibers.
The 3D gel itself is capable of sequestering biomolecules and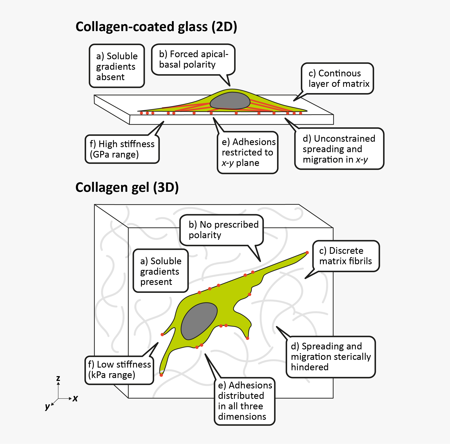 maintaining concentration gradients of soluble factors. For example, the protein aggregates of late-stage neurodegenerative disease are more apparent in 3D than traditional cell culture, where secreted proteins diffuse freely into the culture medium rather than accumulate near cells5,6. In addition, many growth factors are known to bind to ECM proteins. These sequestered growth factors can create concentration gradients that guide stem cell differentiation and morphogenesis during development in vivo7. By contrast, the liquid medium in traditional 2D cell culture does not allow for the storage and release of growth factors and concentration gradients.
maintaining concentration gradients of soluble factors. For example, the protein aggregates of late-stage neurodegenerative disease are more apparent in 3D than traditional cell culture, where secreted proteins diffuse freely into the culture medium rather than accumulate near cells5,6. In addition, many growth factors are known to bind to ECM proteins. These sequestered growth factors can create concentration gradients that guide stem cell differentiation and morphogenesis during development in vivo7. By contrast, the liquid medium in traditional 2D cell culture does not allow for the storage and release of growth factors and concentration gradients.
Finally and most visibly, cells in 3D culture can create 3D structures, rather than being constrained to a single layer in 2D. Many outcomes in morphogenesis and development are three-dimensional, like branches, hollow spheres, tubes, and vascular networks. In multicellular aggregates, the increased dimensionality also allows for increased cell-cell interactions. The functional consequences are evident in cell-dense systems like the liver, where interactions with non-parenchymal cells contribute significantly to hepatocyte function8, and muscle, where spontaneous contractions can be coordinated across all the cells in a microtissue9. Spheroids are a scaffold-free variant of 3D cell culture where aggregated cells may attach to one another and produce their own matrix, reminiscent of the dense organization of solid tumors. Much like solid tumors, tumor spheroids can develop spatial heterogeneity, especially gradients of nutrients and oxygen that alter cell metabolism and susceptibility to anticancer drug10.
3D Cell Culture Advantages
While more technically challenging than traditional cell culture, 3D cell culture supports tissue-specific function and physiological cell-cell and cell-matrix interactions. These factors can contribute to a number of advantages and unique opportunities for 3D cell culture:
- Increased viability of difficult-to-culture cells, including primary cells and prostate tumors11.
- Improved cell-type-specific function and gene expression in cells such as chondrocytes, hepatocytes, myocytes, neurons, and mammary and kidney epithelial cells1,6,9,12–14. For example, features of hepatocytes known to be relevant to drug-induced liver toxicity are more highly expressed in 3D cell culture than in traditional 2D culture.
- Accumulation of cell-secreted proteins in the ECM may have pathological consequences in neurodegenerative disease, and cannot be studied in conventional 2D cell culture. In human stemcell-derived neurons with familial Alzheimer’s, 3D cell culture (but not 2D or mouse models) promoted tauopathy through the accumulation of β-amyloid aggregates in the ECM6. Neural spheroids, but not traditional 2D culture, accumulated amyloid aggregates and tauopathy5.
- And, of course, cells can only produce 3D structures in 3D cell culture. For example, the spontaneous generation of organoids with in vivo-like tissue architecture1, blood vessel formation, branching morphogenesis, and lumenization all require the freedom for cells to self-assemble in 3D, rather than being constrained to a 2D surface.
3D Cell Culture Disadvantages and Limitations
3D cell culture is limited by cost and ease of use:
- ECM proteins or synthetic hydrogels are an additional cost and take some handling skill.
- Fully embedded structures do not permit separate access to apical and basal compartments.
- Lower throughput
- Thick 3D cultures can be difficult to analyze by microscopy:
- Light scatters in many ECM gels, and cell-dense 3D structures may be particularly opaque.
- Large or deeply embedded structures (over 200 microns) may be particularly difficult to image, limited by the working distance of the objective.
- Structures embedded randomly in 3D will not lie at the same focal plane, unlike 2D cell monolayers, making automated microscopy more challenging.
- Collecting cells or secreted factors for biochemical assays is also more challenging in certain ECM gels, such as collagen, which require protease to dissolve and dissociate the embedded cells. On the other hand, some synthetic hydrogels have been specifically designed to dissolve on demand15.
- The investigational nature of many published 3D cell culture systems can result in complex, difficult-to-replicate systems. While great for demonstrating specific organotypic features of 3D cell culture, it’s not easy for other researchers to (for example) recreate a custom-built 3D bioprinter, microfabrication lab, and/or perfusion bioreactor.
Applications of 3D Cell Culture
Unsurprisingly, 3D cell culture is great for studying phenotypes that are poorly reproduced in conventional 2D cell culture, such as amyloid accumulation, chondrogenesis, angiogenesis, cancer metastasis, and morphogenesis. Many of these processes involve cellular behaviors that are intrinsically tied to cell-matrix interactions, whether through synthesis, degradation, directed migration, or mechanical cues.
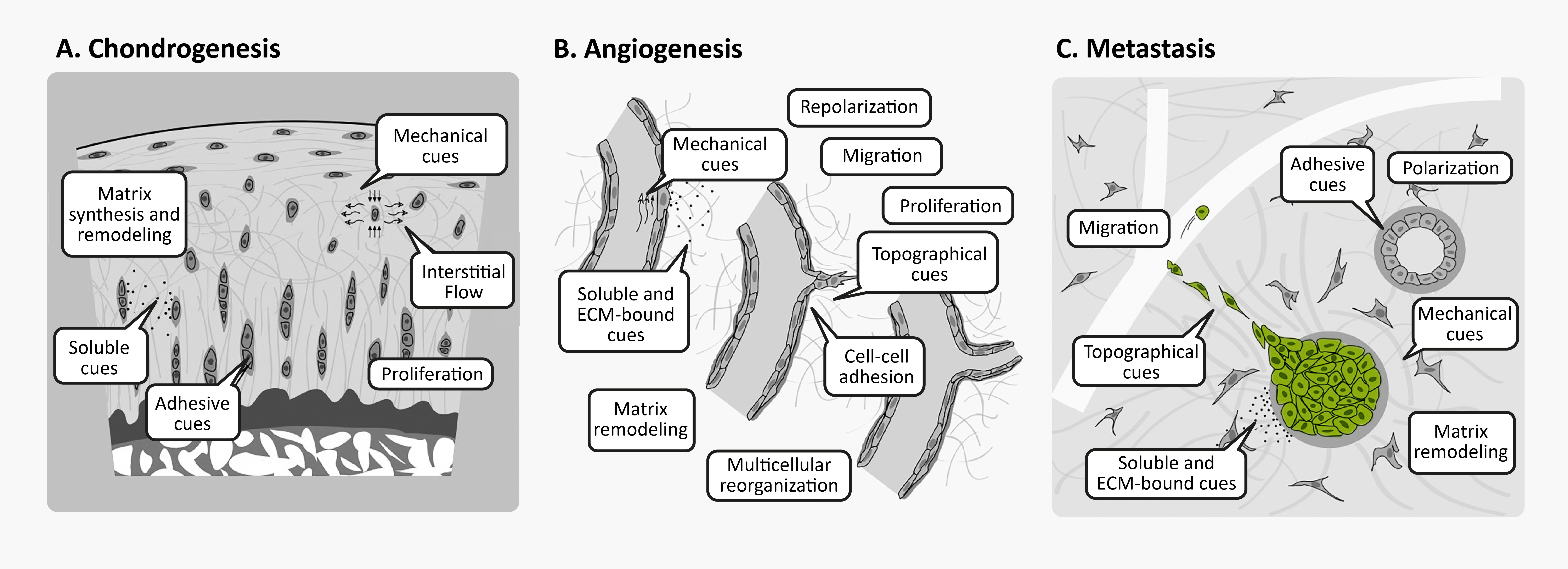
3D cellular phenomena in development tissue homeostasis and disease. Figures from Baker & Chen16
Similarly, 3D morphogenesis in organoids is unlike anything in conventional 2D cell culture. Unlike differentiation of a 2D monolayer of stem cells, 3D aggregates of stem cells exposed to differentiation factors and basement membrane ECM proteins can spontaneously self-organize into tissue structures; in this case, optic cups. Without the constraint of a stiff, 2D substrate, the organoids underwent mechanically driven tissue deformations, just as they do during development17.
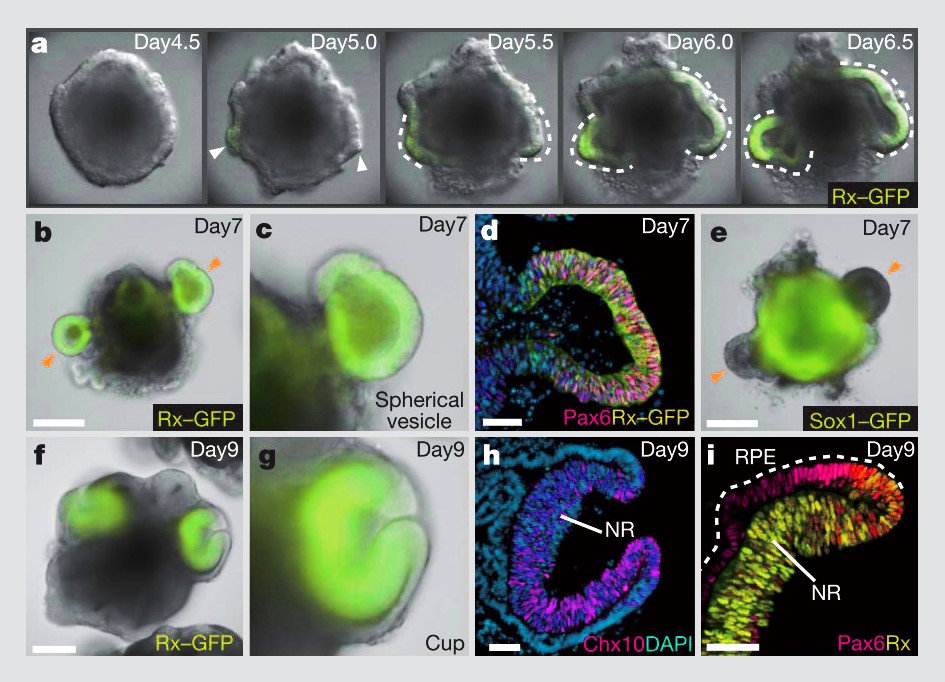
Self formation of an optic cup like structure in 3D culture. Figure from Eiraku et al17.
3D Culture Systems and Methods
3D cell culture is highly customizable, as it should be – every cell type in vivo experiences a different set of chemical and mechanical signals, and pursuing greater physiological relevance requires cell type-specific modifications to cell culture.
The defining choice in 3D cell culture is the type of ECM, which might be any combination of18:
Artificial ECM (including synthetic polymers, self-assembling peptides, and naturally derived materials like silk, cellulose, and alginate)
- Can be engineered to have a range of properties, such as cell degradability, stiffness, porosity, viscosity, texture, and temperature sensitivity
- Compared to animal sources, reduced lot variability and reduced possibility of contamination
- No endogenous cell adhesion sites – must be combined with adhesion proteins or peptides for cells to adhere.
- Minimalist adhesion peptides may not replicate the effect of ECM proteins in vivo, particularly protein-specific effects19.
Natural ECM (ECM proteins like collagen, laminin, and hyaluronic acid, generally purified from animals)
- Most similar to the in vivo environment:
- Extract ECM proteins from specific tissues, which can improve cell differentiation and function20
- Specific ECM proteins for tissue-specific compositions, e.g. more basement membrane-like or more stromal-like.
- Contains native functional sites7 for:
- protease degradation
- growth-factor binding
- cell adhesion, including strain-unfolding adhesion sites
- Purified proteins that are recombined in vitro may not represent the native ECM in composition or nanoscale structure21
- Animal-derived proteins may suffer from lot-to-lot variability and potential contamination
None (scaffold-free, 3D spheroids or microtissues)
- Easier to access and manipulate cells, particularly for high-throughput analysis
- Cells often produce their own matrix over time
- Most similar to traditional 2D cell culture and compatible with existing workflows, such as fluorescence-activated cell sorting22
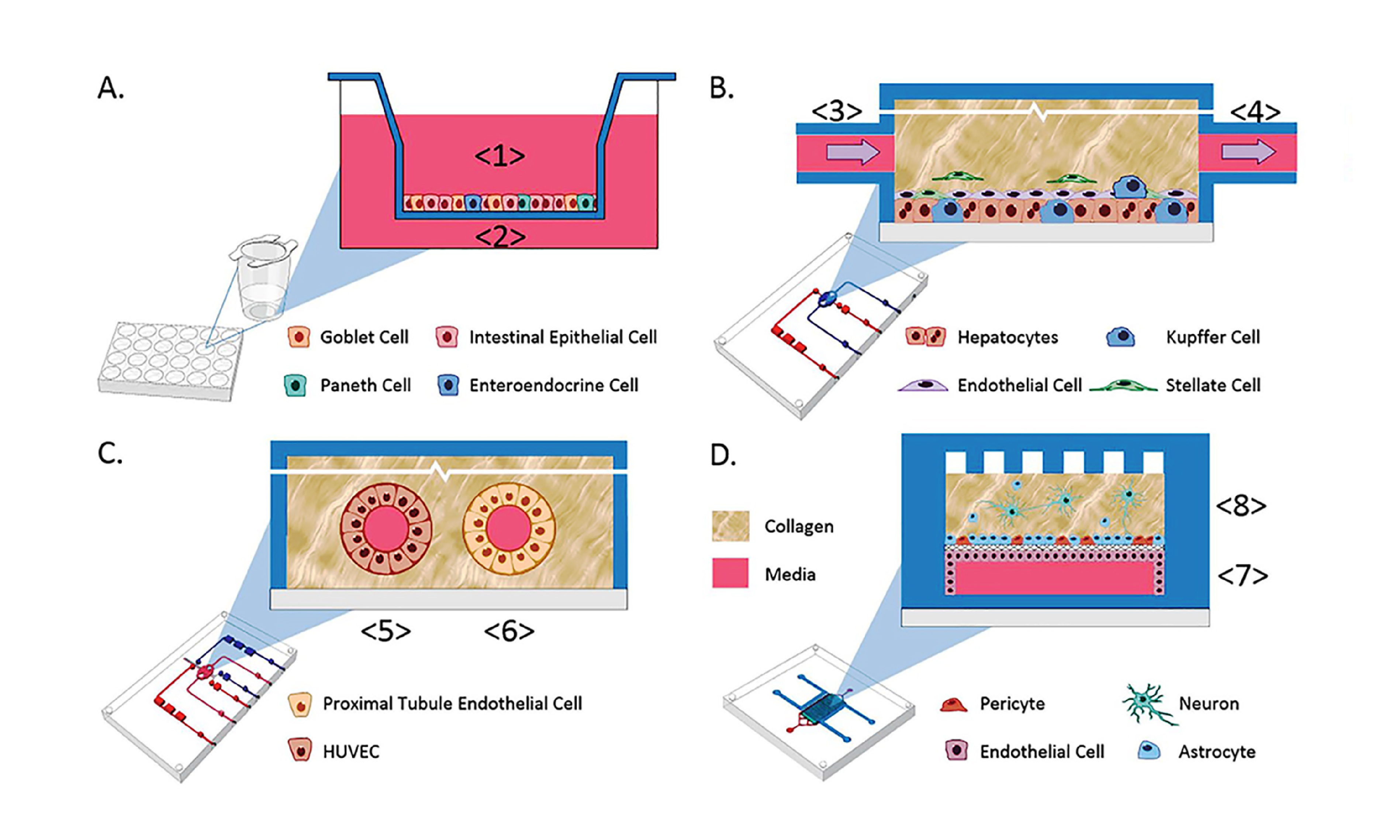
Microfluidic 3D Cell Culture: Organs-on-a-Chip
Besides choice of ECM, you might pursue other tissue-specific choices for recreating an organ in a dish, such as spatial control over the placement of cells, ECM, and soluble factors; different cell types; and fluid transport between compartments. Many 3D cell culture systems are combined with microfluidics, which can provide:
- Easy access to cell-conditioned medium on both the apical and basal sides
- Perfusion for nutrient and oxygen delivery to dense 3D structures
- Controlled exchange and collection of conditioned media between different cell types or different tissue compartments
- Defined concentration gradients of soluble factors
- Spatial control of the shape and interconnectivity of cell compartments
- Low culture volumes can concentrate cell-derived factors and enhance paracrine signaling23
- Mechanical stimuli (interstitial fluid flow, stretch, and fluid shear stress)
- Microfluidic organ-on-a-chip systems frequently incorporate elements of 3D culture into their microfluidic designs, including using natural ECM as a substrate and filling microfluidic chambers with 3D gels. Mechanical cues and spatially separated apical and basal (or lumenal and stromal/vascular) compartments in these systems can offer a richer, more organotypic cell culture system.
In the case of gut-on-a-chip, Caco-2 cells exposed to fluid flow and stretch, but not Caco-2 cells in static membrane culture, displayed villus-like differentiation and morphology and had increased mucus secretion, glucose uptake, and drug metabolism24.
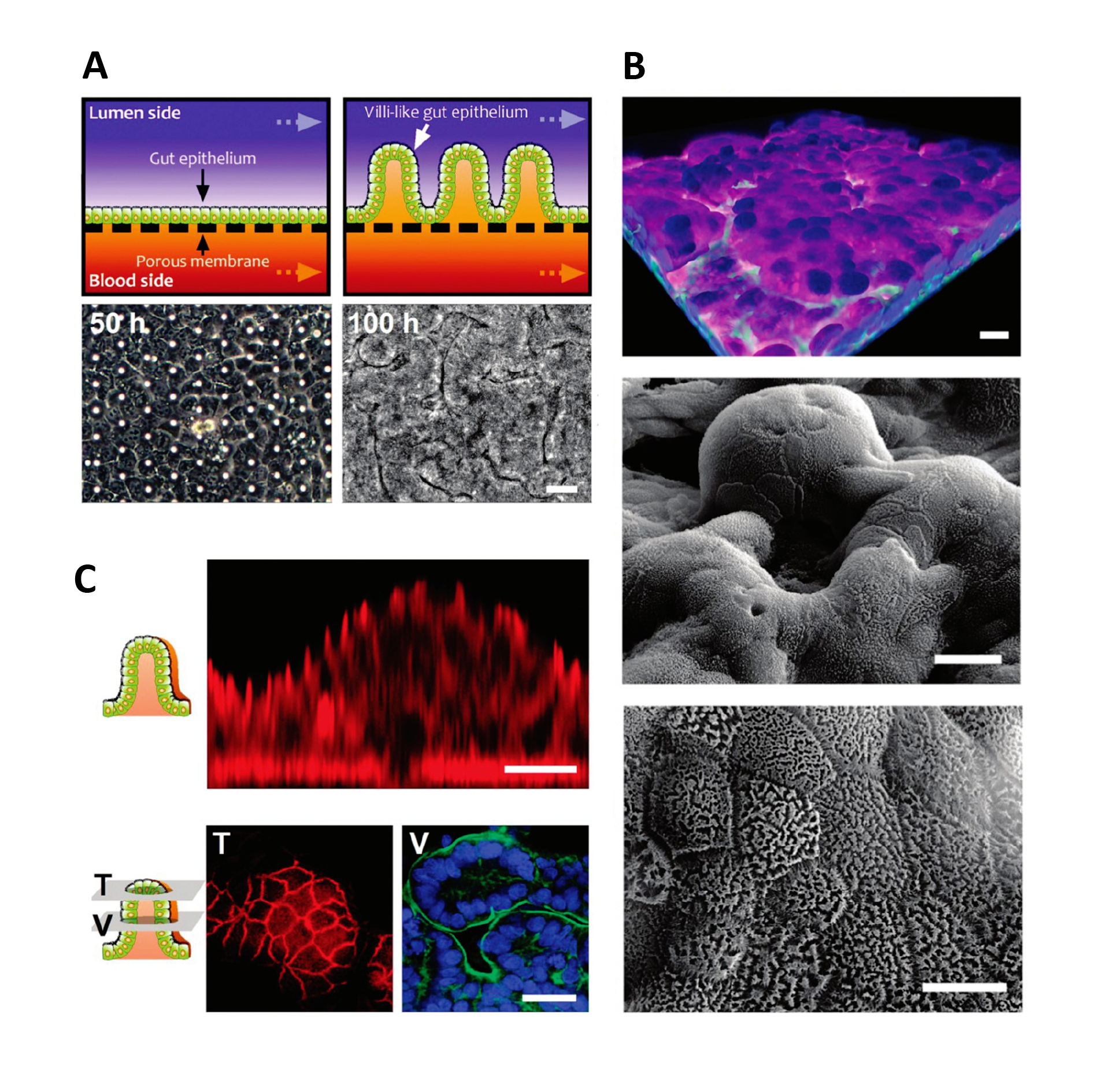
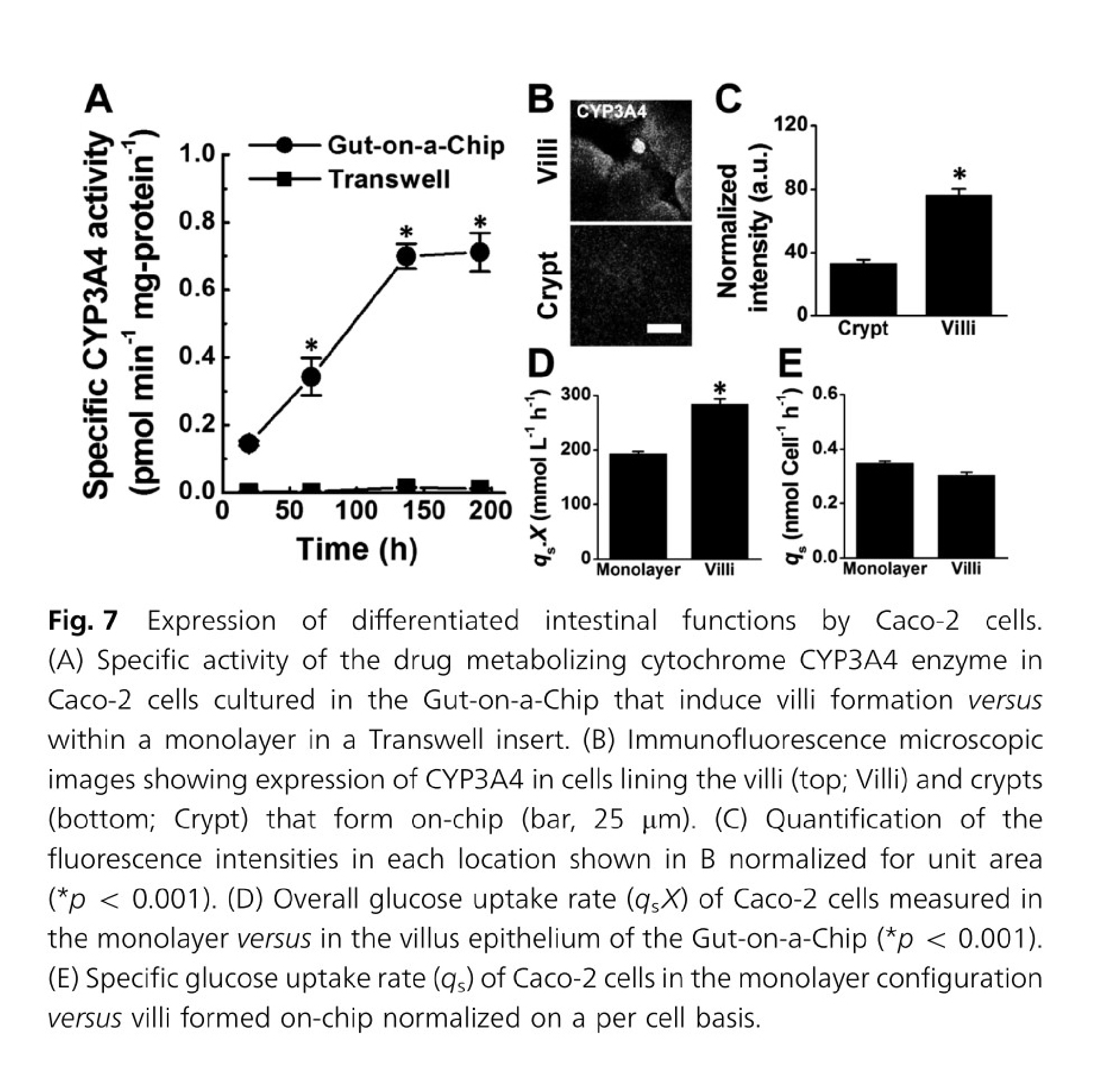
Formation of intestinal villi by Caco 2 cells within in Gut on a Chip cultures. Figures from Kim & Ingber24.
Blood vessel biology in particular has benefited greatly from the combination of 3D cell culture with microfluidic organ-on-a-chip technology. For starters, blood vessel formation and maturation are only possible to observe in 3D cell culture. (In 2D culture, endothelial cells appear a little different from other mesenchymal cells.) Only in 3D cell culture do they demonstrate the ability to form branching, lumenized vascular networks. Researchers interested in angiogenesis have combined 3D culture of endothelial cells with microfluidics to great effect, establishing interstitial fluid flow, shear stress, and growth factor gradients as key signals in blood vessel sprouting25. Similar systems have also been used to study blood vessel anastomosis, the blood-brain barrier, and tumor cell extravasation26.
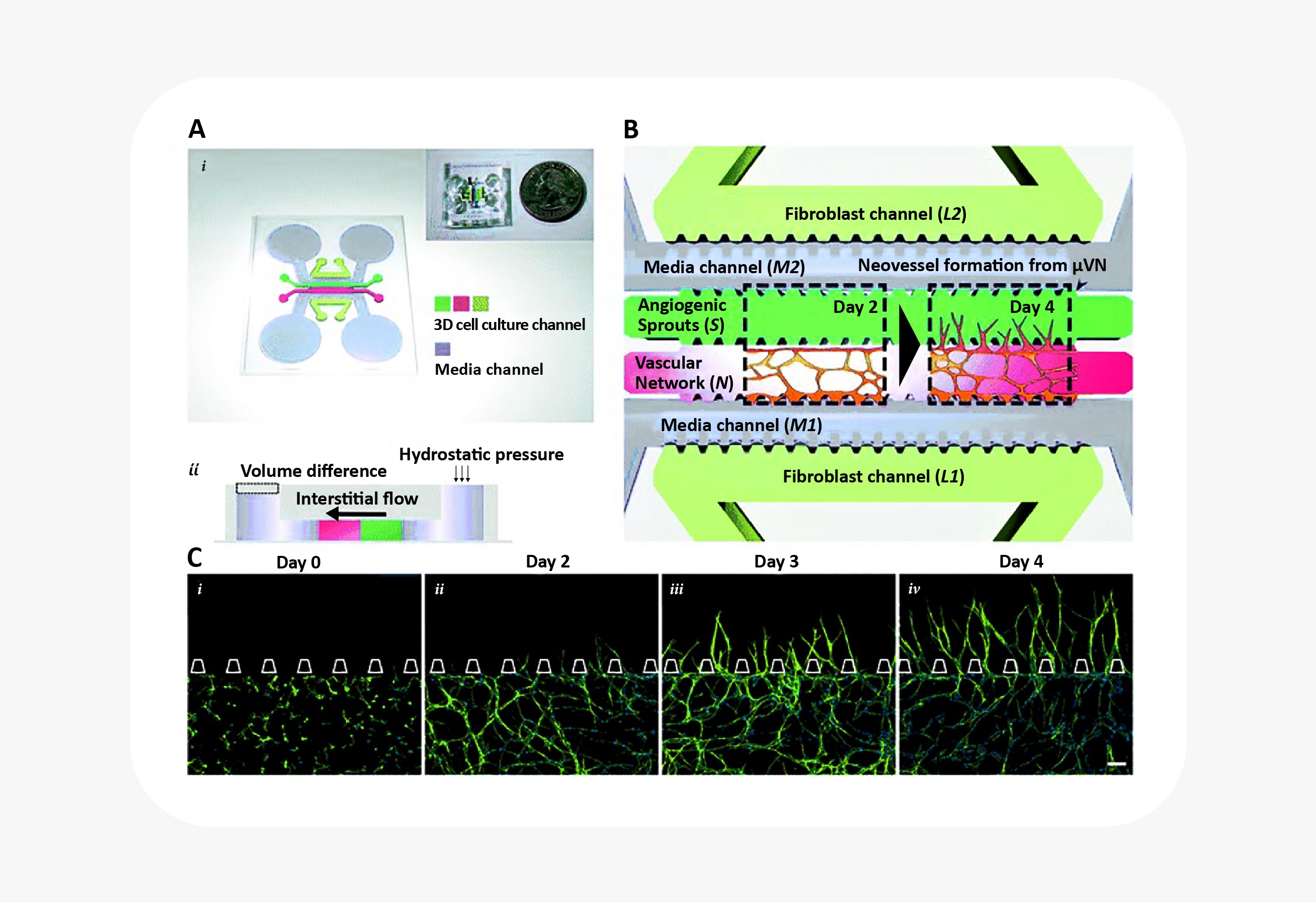
Microfluidic device to reconstitute formation and expansion of microvascular network. Figure from Kim et al25.
Organs-on-a-chip also reflect an appreciation for fluid transport that is often overlooked in other cell culture systems. For example, in a massive feat of coordination, groups across multiple universities worked together to study functional coupling of different microfluidic organs-on-a-chip. They passed conditioned media from one organ model to another in sequence to validate the transport, metabolism, blood-brain barrier permeability, and toxicity of known compounds. Their results not only showed agreement between organ-on-a-chip and clinical data, but also demonstrated the use of sequential fluid transport to study different organs’ effects on downstream organs27.
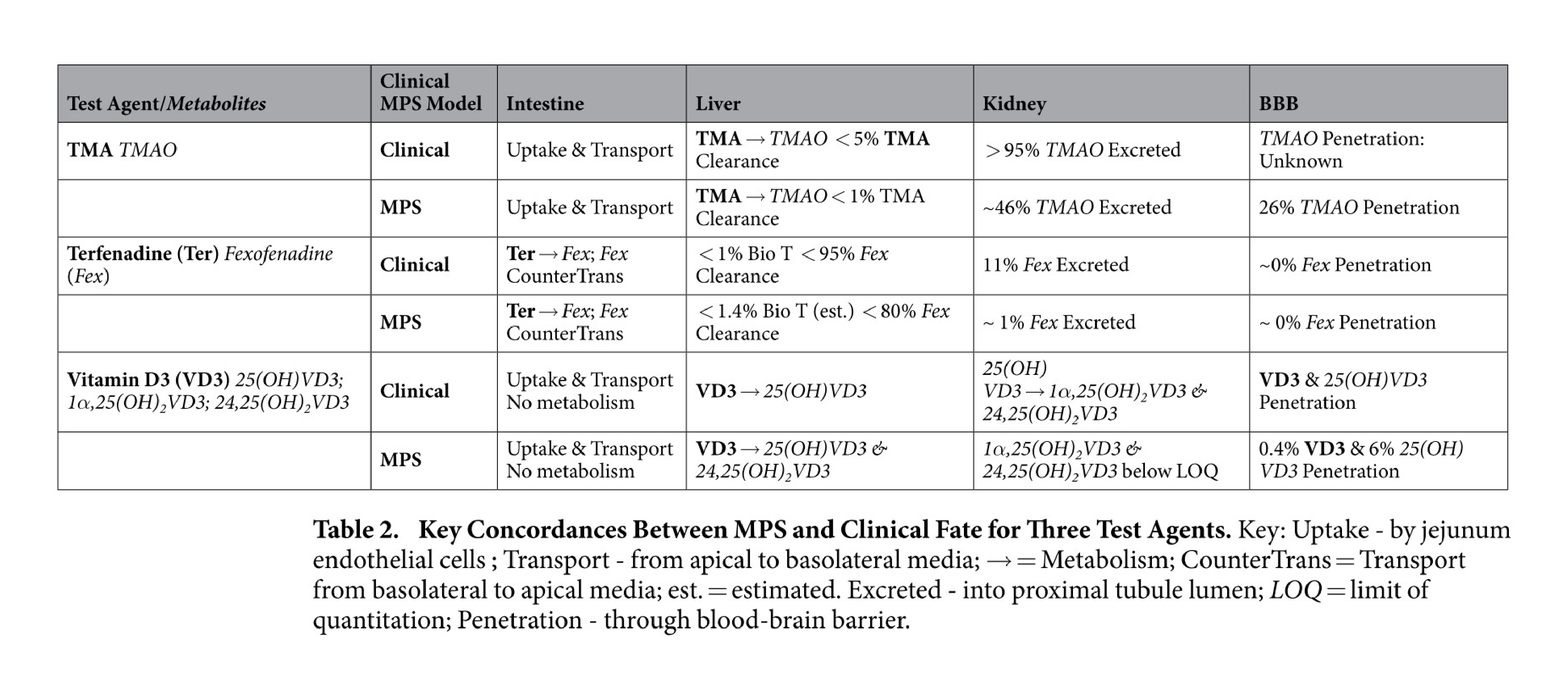
Key concordances between MPS and clinical fate for 3 test agents. Figures from Vernetti et al27
From bench to benches
Of course, these more complex systems may be better models of the body, but the consequence is often more work for the biologist!
Many specialized, investigational 3D culture systems require in-lab microfabrication, syringe pumps to drive fluid flow or pressure, and engineering expertise. Variability in 3D systems between labs makes it hard to generalize results, particularly without a 2D culture internal comparison. 3D assays can be more labor-intensive and, as a result, lower-throughput than possible in 2D culture. Despite the clear advantages of 3D cell culture, adoption by the broader scientific community relies on addressing usability, validation, and researchers’ needs.
As a result, accessible, customizable platforms have been developed to address many common limitations of 3D cell culture:
- Culture chambers of defined, shallow dimensions allow for 3D imaging within the working distance of a microscope objective
- Regularly spaced chambers in multiwell plate format are amenable to automated imaging and compatible with fluid handlers and multi-channel pipettors28
- Standardized, identical channels for reproducibility and high throughput
- Non-destructive live imaging for monitoring function28
- Some devices include integrated sensors, allowing for monitoring of cardiac muscle function29 or barrier permeability30
Notably, the OrganoPlate integrates microfluidic chambers into a standard well plate format, allows spatial control over multiple ECM and cell components, and can be customized to the tissue system and biology of interest. It is a flexible platform that allows the researcher to choose the variables that fit their assay: ECM, cell type(s), co-culture, and perfusion.
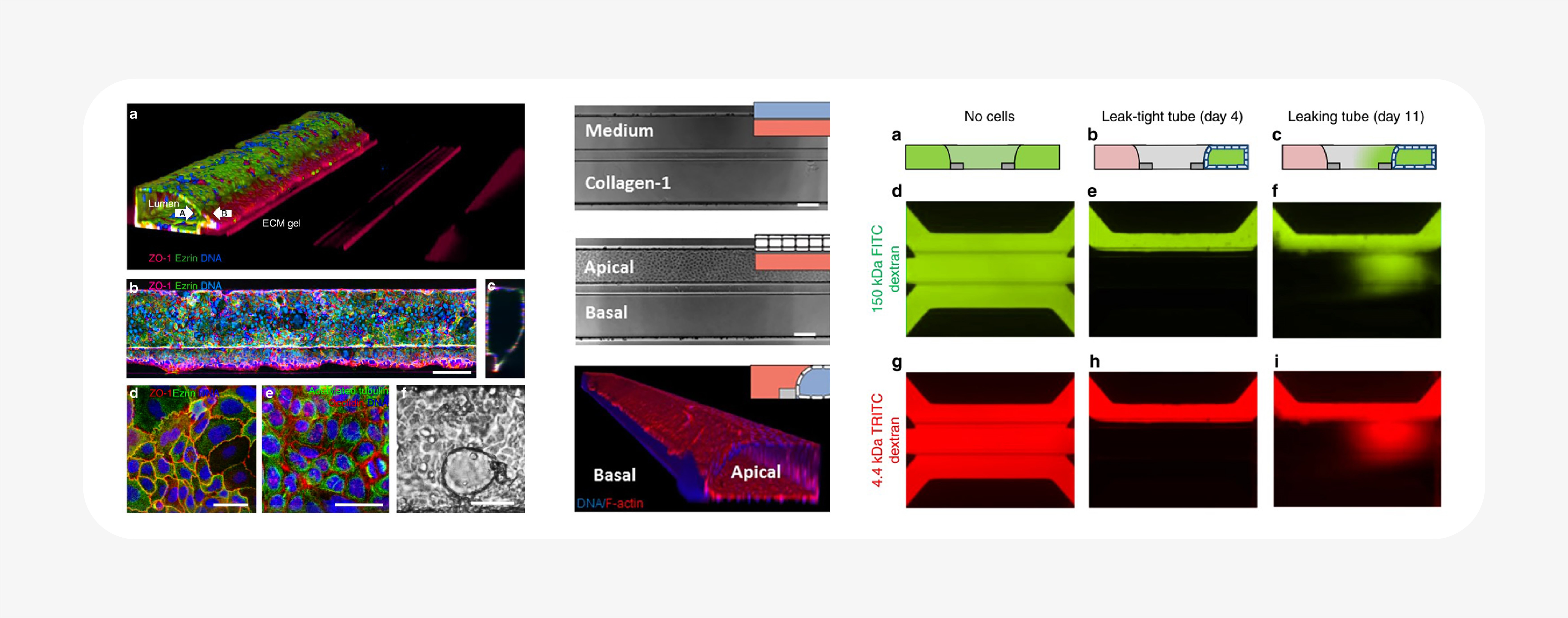
As a result, hundreds of individual ECM-containing tissue cultures can be assayed in parallel. Barrier integrity was reported from a screen of compounds in 357 different intestinal tubes using a fluorescent dextran-based assay28. The same technology was also used to create a high-throughput vascular permeability assay: a plate of 96 perfused, flow-exposed blood vessel tubes, demonstrating its customizability31.
Similarly, while the supporting ECM gel was cell-free in these publications, it would be simple to include stromal or immune cells in the ECM, components of interest in cancer, and stem cell biology. Different drugs or microbiota could be applied to the apical lumens. The possibilities are only limited by the creativity of the researchers! It’s a system that, with remarkably little fuss, can be used to study quite complex in vitro tissues. Even as it incorporates many of the advantages and capabilities of 3D cell culture and microfluidics, it’s also designed to sidestep common throughput and usability downsides.
Get started with 3D cell culture in your lab right now...
Our TechnologyKnowledge Center
References
- Simian, M. & Bissell, M. J. Organoids : A historical perspective of thinking in three dimensions. J. Cell Biol. 216, 1–10 (2016).
- Benam, K. H. et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13, 151–157 (2015).
- Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix Elasticity Directs Stem Cell Lineage Specification. J. Biomech. Eng. 126, 677–689 (2006).
- Baker, B. M. & Chen, C. S. Deconstructing the third dimension – how 3D culture microenvironments alter cellular cues. J. Cell Sci. 125, 3015–3024 (2012).
- Raja, W. K. et al. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One 11, e0161969 (2016).
- Choi, S. H. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
- Muncie, J. M. & Weaver, V. M. The Physical and Biochemical Properties of the Extracellular Matrix Regulate Cell Fate. Current Topics in Developmental Biology 130, (Elsevier Inc., 2018).
- Khetani, S. R. et al. Microengineered Liver Tissues for Drug Testing. J. Lab. Autom. 20, 216–250 (2015).
- Madden, L., Juhas, M., Kraus, W. E., Truskey, G. A. & Bursac, N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife 4, e04885 (2015).
- Hirschhaeuser, F. et al. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 148, 3–15 (2010).
- Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
- Caron, M. M. J. et al. Redifferentiation of dedifferentiated human articular chondrocytes: Comparison of 2D and 3D cultures. Osteoarthr. Cartil. 20, 1170–1178 (2012).
- Ben-Ze’ev, A., Robinson, G. S., Bucher, N. L. R. & Farmer, S. R. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc. Natl. Acad. Sci. 85, 2161–2165 (1988).
- Astashkina, A. I., Mann, B. K., Prestwich, G. D. & Grainger, D. W. Comparing predictive drug nephrotoxicity biomarkers in kidney 3-D primary organoid culture and immortalized cell lines. Biomaterials 33, 4712–4721 (2012).
- Valdez, J. et al. On-demand dissolution of modular, synthetic extracellular matrix reveals local epithelial-stromal communication networks. Biomaterials 130, 90–103 (2017).
- Baker, B. M. & Chen, C. S. Deconstructing the third dimension - how 3D culture microenvironments alter cellular cues. J. Cell Sci. 125, 3015–3024 (2012).
- Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011).
- Astashkina, A., Mann, B. & Grainger, D. W. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol. Ther. 134, 82–106 (2012).
- Yu, W. B1-Integrin Orients Epithelial Polarity via Rac1 and Laminin. Mol. Biol. Cell 16, 433–445 (2004).
- Zhang, Y. et al. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials 30, 4021–4028 (2009).
- Halfter, W. et al. New concepts in basement membrane biology. FEBS J. 282, 4466–4479 (2015).
- Birchler, A. et al. Seamless Combination of Fluorescence-Activated Cell Sorting and Hanging-Drop Networks for Individual Handling and Culturing of Stem Cells and Microtissue Spheroids. Anal. Chem. 88, 1222–1229 (2016).
- Domenech, M. et al. Cellular observations enabled by microculture: Paracrine signaling and population demographics. Integr. Biol. 1, 267–274 (2009).
- Kim, H. J. & Ingber, D. E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. (United Kingdom) 5, 1130–1140 (2013).
- Kim, S., Chung, M., Ahn, J., Lee, S. & Jeon, N. L. Interstitial flow regulates the angiogenic response and phenotype of endothelial cells in a 3D culture model. Lab Chip 16, 4189–4199 (2016).
- Bersini, S., Jeon, J. S., Moretti, M. & Kamm, R. D. In vitro models of the metastatic cascade: From local invasion to extravasation. Drug Discov. Today 19, 735–742 (2014).
- Vernetti, L. et al. Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle. Sci. Rep. 7, 42296 (2017).
- Trietsch, S. J. et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 8, 1–7 (2017).
- Lind, J. U. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 16, 303–308 (2017).
- Henry, O. Y. F. et al. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 17, 2264–2271 (2017).
- Van Duinen, V. et al. 96 Perfusable Blood Vessels To Study Vascular Permeability in Vitro. Sci. Rep. 7, 1–11 (2017).
Copyright to every image belongs to the original source, linked as a reference in each text in or around the image.
